NIHilist's Immunology
How Immune System works
Wednesday, January 11, 2023
If T cell clones are so diverse, what prevents anti-tumor immune response?
Friday, September 18, 2020
A specific bacteria-infecting virus, bacteriophage, found in gut microflora, augments anti-tumor T cell immunity
Molecular mimicry between microbial and host's antigens could contribute to autoimmunity but also to the protection against tumors through epitope cross-reactivity. A new study in journal Science indicates that those cross-reactive epitopes could come from viruses that infect endogenous microbial species.
In this study the authors made a surprising observation that only certain Enterococcus hirae microbial strains (E. hirae 13144 or IGR11) augmented anti-cancer effect in experimental cancer model.
Next, the authors showed that this biological activity was linked to one dominant epitope, TSLARFANI, derived from TMP protein that originated in 39.2-kb prophage only in those specific E. hirae strains. Mice immunized with heat-inactivated E. hirae 13144 strain, or peptide TSLARFANI, or irrelevant E.coli engineered to express TMP, all augmented anti-cancer effect.
Mechanistically, the authors showed that epitope, GSLARFRNI, derived from cancer cells used in these experiments, was recognized by the same CD8 T cells which labeled with TSLARFANI epitope tetramers confirming cross-reactivity between these 2 epitopes.
In summary, this study suggests that microbiota and bacteriophages they carry represent new modality in fight against cancer. In this study overall anti-tumor effect is modest but we need to take into account that this is an effect of just one cross-reactive epitope in one type of MHC inbred mice, and it is likely that many other epitopes will be involved in outbred species such as humans. However, it is still extremely hard to do such analysis in humans in real world scenario due to lack of exact knowledge about human microbiota strains and poor reliability and performances of available bioinformatics approaches. However, once the mechanistic principles underlying anti-cancer effects are uncovered and accepted, then it is much easier to move the field forward.
posted by David Usharauli
Thursday, August 6, 2020
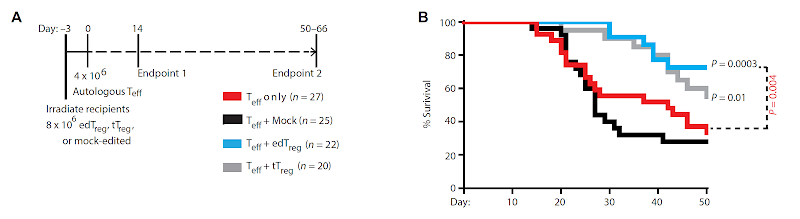
Is it possible to engineer Foxp3+ Tregs from primary T cells?
 |
 |
This
could also explain why the authors did not see improvement in brain
inflammation in mice EAE model when co-transferring antigen-specific edTregs with
effector T cells.  |